Lead: Scott Ritchie
A major research question of the upcoming precision medicine era is: can we utilise large-scale omics data to infer the biological processes underlying new biomarker associations?
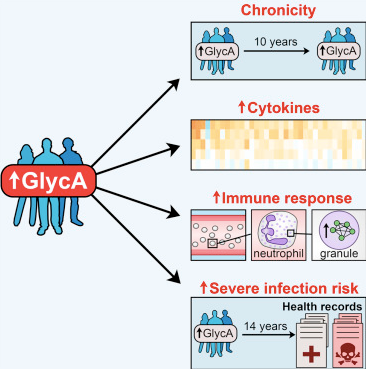 We are investigating various strategies to answer this question and have recently published a study which identifies chronic low-grade inflammation, potentially a neutrophilic response to microbial insults, as a putative explanation for the GlycA biomarker's association with hospitalisation and mortality, particularly from infections (Ritchie et al. 2015). We are furthering our exploration of GlycA's relationship with incident cardiovascular and other diseases, as well as fine-mapping the biomarker to identify specific glycoproteins that may themselves exhibit causal effects.
We are investigating various strategies to answer this question and have recently published a study which identifies chronic low-grade inflammation, potentially a neutrophilic response to microbial insults, as a putative explanation for the GlycA biomarker's association with hospitalisation and mortality, particularly from infections (Ritchie et al. 2015). We are furthering our exploration of GlycA's relationship with incident cardiovascular and other diseases, as well as fine-mapping the biomarker to identify specific glycoproteins that may themselves exhibit causal effects.
Additional studies include:
- Creating integrative maps of immunometabolic interactions and their genomic regulation.
- Identifying the pathways through which polygenic risk of disease operate.
- Methodologies for the integrative analysis of multi-omics data and prospective health records.
Relevant reading
A scalable permutation approach reveals replication and preservation patterns of network modules in large datasets
Cell Systems 2016 3(1):71–82
The biomarker GlycA is associated with chronic inflammation and predicts long-term risk of severe infection
Cell Systems 2015 1(4):293–301
Novel loci for metabolic networks and multi-tissue expression studies reveal genes for atherosclerosis
PLOS Genetics 2012 8(8):e1002907