14 September 2020
Institute news
Melbourne researchers have developed a tool to monitor mutations that make it difficult to develop coronavirus (COVID-19) vaccines and drugs.
Ensuring treatments remain effective as the virus mutates is a huge challenge for researchers. The powerful new tool harnesses genomic and protein information about the virus and its mutations to aid drug and vaccine development.
 The Baker Institute’s Associate Professor David Ascher and his colleagues at the Bio21 Molecular Science and Biotechnology Institute and the University of Melbourne developed the software tool and library, dubbed COVID-3D.
The Baker Institute’s Associate Professor David Ascher and his colleagues at the Bio21 Molecular Science and Biotechnology Institute and the University of Melbourne developed the software tool and library, dubbed COVID-3D.
This advance builds on the Institute’s growing work in the area of bioinformatics, with our computational scientists harnessing big data to accelerate and target disease prediction, prevention and treatment.
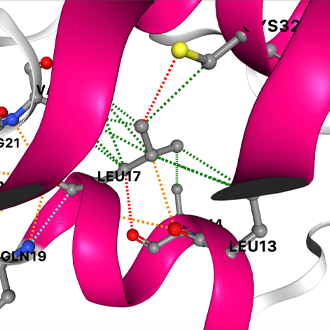
Published in Nature Genetics, COVID-3D contains information about all the protein structures that coincide with the SARS-CoV-2 (COVID-19) genome, including every known genetic mutation and its resultant mutant protein structure.
“Although the SARS-CoV-2 virus is a relatively new pathogen, its ability to readily accumulate mutations across its genes was evident from the start of this pandemic,” Associate Professor Ascher said.
“In the context of therapeutic drug design and discovery, these mutations, and the patterns by which they accumulate within the virus’ protein structures, can affect the ability of vaccines and drugs to bind the virus, or to create a specific immune response against it. Because of this, scientists must not only try to control the virus, but outsmart it by predicting how it will change over time.”
To develop COVID-3D, Professor Ascher’s team analysed the genome sequencing data of over 120,000 SARS-CoV-2 samples from infected people globally, including those that uniquely affect Australia, to identify mutations within each of the virus’ proteins. They tested and analysed the mutations’ effects on their protein structure using computer simulations.
This data was used to calculate all the biological effects of every possible mutation within the genome.
Mutations or changes in an organism’s genetic material are natural ‘errors’ in the cell replication process. They can give the virus new ‘powers’ of survival, infectivity and virulence. Fortunately, the researchers found SARS-CoV-2 is mutating slower than other viruses such as influenza, with about two new changes in its genome every month.
COVID-3D can help researchers recognise how mutations operate and identify more effective vaccine and drug targets. “It is only when you know how a mutation will affect the 3D shape of a protein, that you can predict if it will compromise your drug’s ability to bind,” Associate Professor Ascher said.
“As the global scientific and medical community gains better understanding of the biology behind the SARS-CoV2 infection and disease, this will be a powerful resource to predict problems with mutations and to guide the development of more effective therapies.”
Stay on top of the latest findings with our newsletter
Subscribe




